Research by Antonis Klonizakis et al. at the Nikolaou lab uncovers a hidden layer of complexity in mammalian gene regulation and hints at the existence of "negative enhancer" elements.
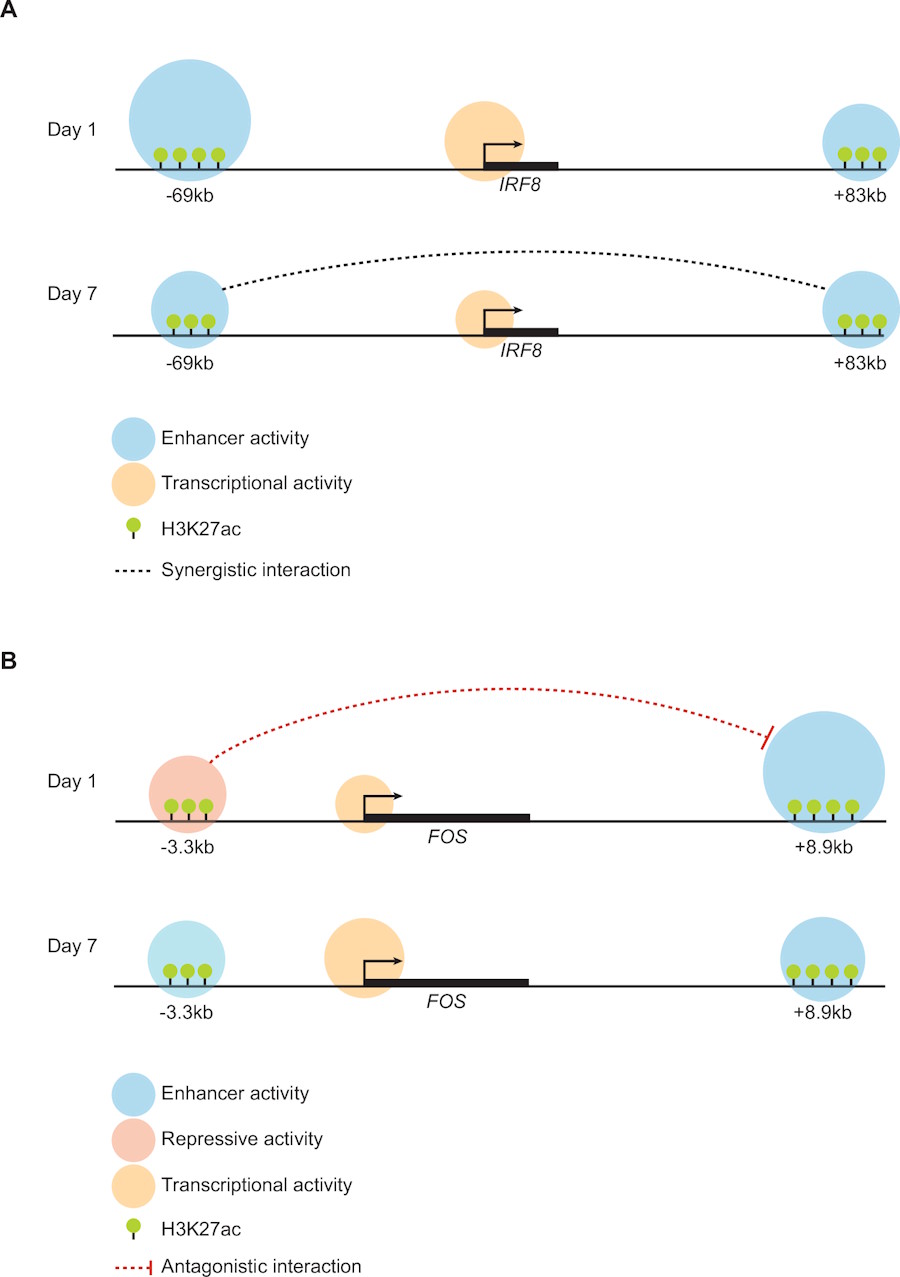
One of the most interesting questions in biology is how the same genome gives rise to a multitude of different cell types. Cell-fate determining genes are usually regulated by enhancer clusters linked to very high expression of nearbly genes through the formation of transcriptional condensates. In this work, in collaboration with the lab of Thomas Graf at CRG, Barcelona, we deployed a systems biology approach to explicitly dissect the regulatory "grammar" of super-enhancers in a model of b-cells transdifferentiation to macrophages. After applying SEGCOND, our own computational method for identifying transcriptional condensates (https://pubmed.ncbi.nlm.nih.gov/36394233/) we selected two super-enhancers near the IRF8 and FOS loci respectively and went on to selectively remove (with Crispr-Cas9) combinations of their constituent regulatory elements. We then used statistical modeling to compare the modified enhancers with the resulting gene expression levels.
In the case of IRF8 we found two distinct enhancer elements to be working in synergy but with one having a much stronger effect compared to the other. Using a deep-learning approach to analyze the impact of their underlying DNA sequences we could identify the regulatory contributions to two different transcriptional regulators, PU.1 and CEBPA.
What we found in the case of FOS was rather unexpected. Here, we were also able to identify two distinct enhancer elements, which however showed a strong antagonistic relationship, with one functioning as a strong PU.1/CEBPA activator and the other functioning as a platform for the ETV6 transcriptional repressor. Interestingly, this antagonistic interaction was sufficient to fine-tune FOS expression in a time-dependent manner. Under the influence of the ETV6 repressor, FOS was under-expressed at the early stages of transdifferentiation, but was up-regulated at a later time-point, when ETV6 itself became down-regulated.
While these two cases were dissected in detail experimentally, numerous others have also been identified through computational predictions. Our work strongly suggests the existence of an intricate (and as yet overlooked) regulatory grammar, whereby enhancer elements can combine synergistically and even antagonistically in a way that may give rise to equally sophisticated gene expression programs.
Klonizakis A, Alcoverro-Bertran M, Massó P, Thomas J, de Andrés-Aguayo L, Wei X, Varamogianni-Mamatsi V, Nikolaou C, Graf T. Synergistic and antagonistic activities of IRF8 and FOS enhancer pairs during an immune-cell fate switch. EMBO J. 2025 Apr;44(7):2025-2055.
doi: 10.1038/s44318-025-00380-w.